Tutorial 1. Python tutorial for quantum chemistry simulation
Learning objectives
- Learn basic python scripting to run a quantum chemistry simulation from scratch
- Topics:
- Geometry generation
- Input file generation
- Output file processing
Prerequisites
- Conda installed on your system
- Create new Conda environment called python_tutorial with the following packages installed: jupyter, numpy, matplotlib
- Terachem installed on a GPU cluster using SGE queue manager
Example Task:
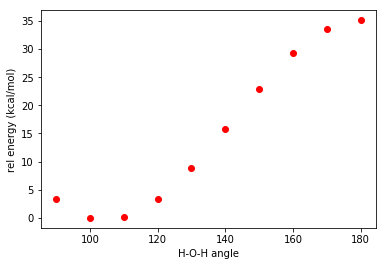
Study the potential energy curve of a water molecule at various H-O-H angles while keep the bond length fixed.
Generate geometries
An .xyz file has the following general format
3
O 0.00000 0.000000 0.000000
H -0.672459 0.672459 0.000000
H 0.672459 0.672459 0.000000Put the water molecule on the x-y plane with O at the origin, it’s easy to write down the expression of the coordinates.
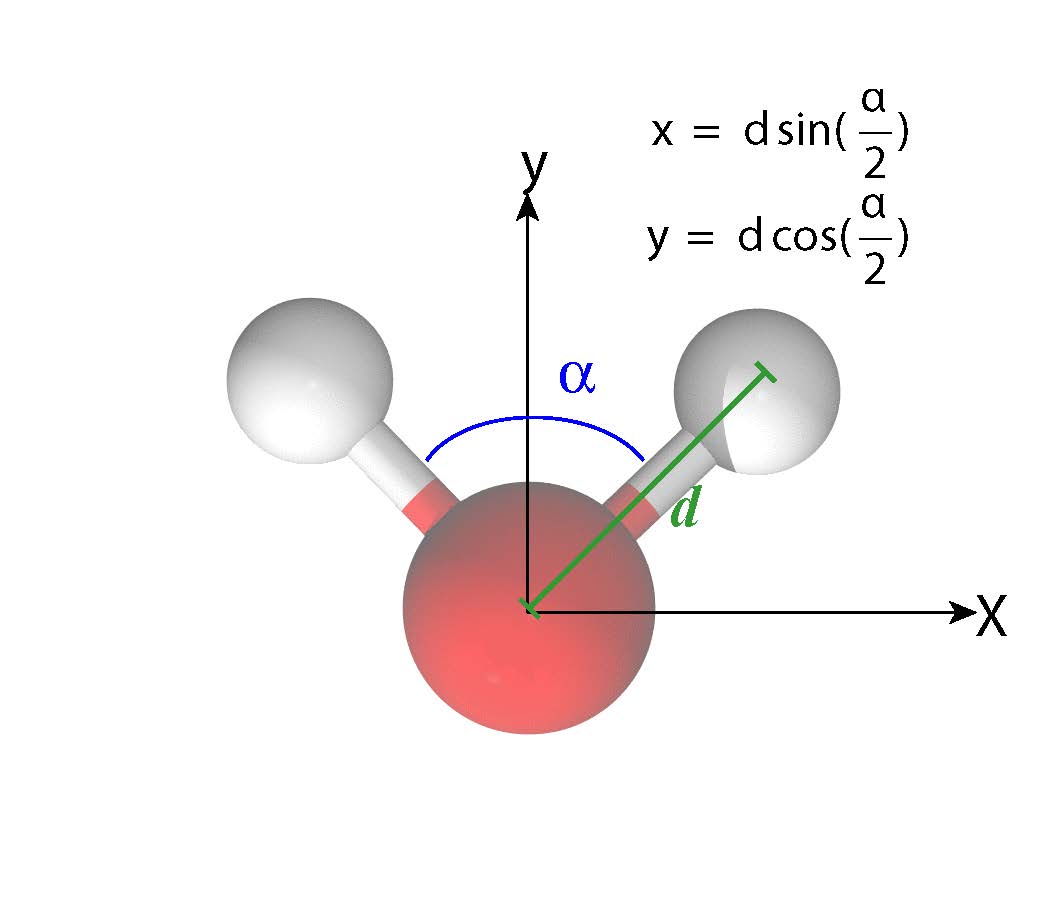
import numpy as np
def writeHOHxyz(bl, angle_min, angle_max, angle_increment):
for deg in np.arange(angle_min, angle_max + 1, angle_increment):
x = np.sin(np.deg2rad(deg / 2.0)) * bl
y = np.cos(np.deg2rad(deg / 2.0)) * bl
xyzname = "HOH-bl-" + str(bl) + "-deg-" + str(deg).zfill(3) + ".xyz"
print(xyzname)
xyz = open(xyzname, "w")
#TODO (Exercise): Write out the lines in xyz file
writeHOHxyz(0.951, 90, 180, 10)TeraChem Input Generation
To run a terachem job on our GPU cluster, one needs at least 3 files: geometry, input deck, submission script.
Here we are going to write a script to generate terachem input deck and submission scripts in batch.
A basic terachem input deck looks like the following:
# TeraChem input deck: energy.in
coordinates HOH-bl-0.951-deg-090.xyz
charge 0
spinmult 1
basis 6-31g*
method b3lyp
run energy
scf diis+a
timings yes
endA basic jobscript for the SGE queueing system looks like the following:
#$ -S /bin/bash
#$ -N HOH-bl-0.951-deg-090
#$ -l h_rt=00:05:00
#$ -l gpus=1
#$ -cwd
#$ -q gpus*
#$ -pe smp 1
# -fin *
# -fout *
export OMP_NUM_THREADS=1
module load terachem/tip
terachem energy.in > energy.outNow let us write a python script that can generate the input file for all the geometries we generated in one batch:
- Create a separate folder for each geometry
- Inside the folder, put in all the needed files: geometry, input deck, and jobscript
- (On the cluster) Job generation and job submission all at once
Basically, the script will create the following tree for us:
.
├── HOH-bl-0.951-deg-090
│ ├── HOH-bl-0.951-deg-090.xyz
│ ├── energy.in
│ └── jobscript
├── HOH-bl-0.951-deg-100
│ ├── HOH-bl-0.951-deg-100.xyz
│ ├── energy.in
│ └── jobscript
├── HOH-bl-0.951-deg-110
│ ├── HOH-bl-0.951-deg-110.xyz
│ ├── energy.in
│ └── jobscript
├── HOH-bl-0.951-deg-120
│ ├── HOH-bl-0.951-deg-120.xyz
│ ├── energy.in
│ └── jobscript
├── HOH-bl-0.951-deg-130
│ ├── HOH-bl-0.951-deg-130.xyz
│ ├── energy.in
│ └── jobscript
├── HOH-bl-0.951-deg-140
│ ├── HOH-bl-0.951-deg-140.xyz
│ ├── energy.in
│ └── jobscript
├── HOH-bl-0.951-deg-150
│ ├── HOH-bl-0.951-deg-150.xyz
│ ├── energy.in
│ └── jobscript
├── HOH-bl-0.951-deg-160
│ ├── HOH-bl-0.951-deg-160.xyz
│ ├── energy.in
│ └── jobscript
├── HOH-bl-0.951-deg-170
│ ├── HOH-bl-0.951-deg-170.xyz
│ ├── energy.in
│ └── jobscript
├── HOH-bl-0.951-deg-180
│ ├── HOH-bl-0.951-deg-180.xyz
│ ├── energy.in
│ └── jobscript
├── jobgen-multi-submission.py
└── xyzfiles
├── HOH-bl-0.951-deg-090.xyz
├── HOH-bl-0.951-deg-100.xyz
├── HOH-bl-0.951-deg-110.xyz
├── HOH-bl-0.951-deg-120.xyz
├── HOH-bl-0.951-deg-130.xyz
├── HOH-bl-0.951-deg-140.xyz
├── HOH-bl-0.951-deg-150.xyz
├── HOH-bl-0.951-deg-160.xyz
├── HOH-bl-0.951-deg-170.xyz
└── HOH-bl-0.951-deg-180.xyzFirst in your terminal, create a new directory. Go to that directory and create a folder called “xyzfiles”, and copy all your generated xyz files into that folder. Run “pwd” and remember the path that is displayed. Then let’s move to that directory in our Jupyter notebook.
import os
my_starting_path=os.path.abspath('.')
print(my_starting_path)
import shutil
import glob
#first write terachem input files
os.chdir(my_starting_path)
xyz_path = os.path.abspath('xyzfiles')
base_submission_path = os.path.abspath(my_starting_path)
os.chdir(xyz_path)
for xyz in glob.glob("*.xyz"):
molecule_name = xyz[:-4]
print(molecule_name)
submission_path = os.path.join(base_submission_path, molecule_name)
if not os.path.isdir(submission_path):
os.makedirs(submission_path)
os.chdir(submission_path)
shutil.copy(
os.path.join(xyz_path, xyz), os.path.join(submission_path, xyz))
tcin = open('energy.in', 'w')
#TODO (Exercise): identify the missing keyword and fill it in here
tcin.write('charge 0\n')
tcin.write('spinmult 1\n')
tcin.write('basis 6-31g*\n')
tcin.write('method b3lyp\n')
tcin.write('run energy\n')
tcin.write('scf diis+a\n')
tcin.write('timings yes\n')
tcin.write('end\n')
tcin.close()
jobscript = open('jobscript', 'w')
jobscript.write('#$ -S /bin/bash\n')
#TODO (Exercise): fill in a line that specify the jobname
jobscript.write('#$ -l h_rt=00:05:00\n')
jobscript.write('#$ -l gpus=1\n')
jobscript.write('#$ -cwd\n')
jobscript.write('#$ -q gpus*\n')
jobscript.write('#$ -pe smp 1\n')
jobscript.write('# -fin *\n')
jobscript.write('# -fout *\n\n')
jobscript.write('export OMP_NUM_THREADS=1\n')
jobscript.write('module load terachem/tip\n')
jobscript.write('terachem energy.in > energy.out\n')
jobscript.close()Now in your terminal, type
import subprocess
subprocess.call('tree')And check what you have. Slight modification of your script can enable batch job submission too!
import os
import shutil
import glob
import subprocess
os.chdir(my_starting_path)
#first write terachem input files
xyz_path=os.path.abspath('xyzfiles')
base_submission_path=os.path.abspath(my_starting_path)
os.chdir(xyz_path)
for xyz in glob.glob("*.xyz"):
molecule_name = xyz[:-4]
print(molecule_name)
submission_path = os.path.join(base_submission_path, molecule_name)
if not os.path.isdir(submission_path):
os.makedirs(submission_path)
os.chdir(submission_path)
shutil.copy(os.path.join(xyz_path,xyz),os.path.join(submission_path,xyz))
tcin = open('energy.in','w')
tcin.write('coordinates %s.xyz\n' %molecule_name)
tcin.write('charge 0\n')
tcin.write('spinmult 1\n')
tcin.write('basis 6-31g*\n')
tcin.write('method b3lyp\n')
tcin.write('run energy\n')
tcin.write('scf diis+a\n')
tcin.write('timings yes\n')
tcin.write('end\n')
tcin.close()
jobname=molecule_name
jobscript = open('jobscript','w')
jobscript.write('#$ -S /bin/bash\n')
jobscript.write('#$ -N %s\n' %jobname)
jobscript.write('#$ -l h_rt=00:05:00\n')
jobscript.write('#$ -l gpus=1\n')
jobscript.write('#$ -cwd\n')
jobscript.write('#$ -q gpus*\n')
jobscript.write('#$ -pe smp 1\n')
jobscript.write('# -fin *\n')
jobscript.write('# -fout *\n\n')
jobscript.write('export OMP_NUM_THREADS=1\n')
jobscript.write('module load terachem/tip\n')
jobscript.write('terachem energy.in > energy.out\n')
jobscript.close()
subprocess.call('qsub jobscript',shell=True)TeraChem Output Processing
Here we’d like to read out the final energy from the terachem output files and convert the unit to kcal/mol, and plot the potential energy curve.
# Please modify the path (add absolute path) if this doesn't work for you
os.chdir(my_starting_path+ "/output_processing")
base_submission_path = os.path.abspath('.')
subdirs = glob.glob(base_submission_path + "/*/")
Energy = dict()
for dir in subdirs:
mydir = dir
if dir[-1] == '/':
mydir = dir[:-1]
output = os.path.join(mydir, "energy.out")
energy = "NA"
if os.path.exists(output):
data = open(output).readlines()
#TODO (Exercise): loop through the lines in output and look for the calculated SCF energy
for line in data:
Emin = min(Energy.values())
HartreeToKcal = 627.509
for key in sorted(Energy):
Energy[key]=(Energy[key] - Emin) * HartreeToKcal
print(key, Energy[key])Plot results
Here we plot with matplotlib, but there are many tools for creating scientific figures. We will cover that topic in future tutorials.
import matplotlib.pyplot as plt
plt.plot(Energy.keys(),Energy.values(),'ro')
plt.ylabel('rel energy (kcal/mol)')
plt.xlabel('H-O-H angle')
plt.show()This will generate the potential energy curve shown at the beginning of this tutorial.
