Tutorial 18. Visualize normal modes/energy gradients/non-adiabatic coupling vector
Learning objectives
- Learn to create static graphs with molecule structure labeled with arrows that represent some interesting vectors, such as normal modes, energy gradients, and even non-adiabatic coupling vectors.
- Learn to create animations of molecular vibration in given the normal mode vector
Background
Normal mode analysis is the essential tool to study polyatomic vibration. The eigenvalues ($\lambda$) obtained from normal mode analysis tell us the vibration frequencies ($\nu$ is related to $\sqrt{\lambda}$). The eigenvectors (the normal mode vectors) are also valuable because they reveal what kind of motion that mode corresponds to. Visualization of the normal modes gives us an intuitive picture of what is happening when that mode vibrates.
For example, one can visualize the normal modes as arrows along with the 3D molecular structure.
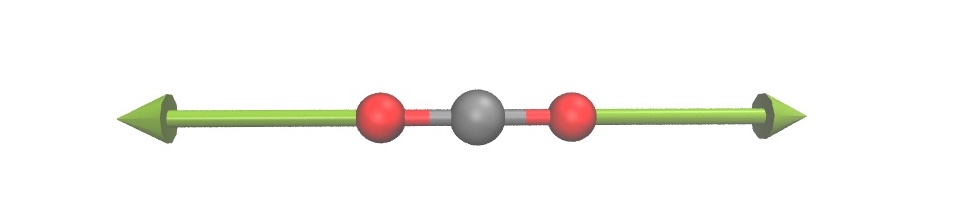
One can also create animations of the vibration of a normal mode.
Apart from the normal modes, there are other vectors we want to visualize in computational chemistry research. For example, the gradients of energy with respect to nuclear coordinates, and non-adiabatic coupling vectors between two electronic states. All these vectors have the following features: have 3N dimensions for an N-atom molecule, and are related to the 3 cartesian coordinates of each atom. You might have seen publications with figures showing a molecule structure with arrows on each atom indicating the normal modes or nonadiabatic coupling vectors. How are those figures generated?
Some commercial quantum chemistry packages compute Hessian matrices at the minimized structure and automatically do normal mode analysis, and generate corresponding file formats to be used in the corresponding commercial visualizers.
However, suppose you don’t have access to the commercial visualizers. You have manually calculated the normal modes of a simple molecule in your textbook exercise or downloaded the normal modes from a database. You know the 3D coordinates of that molecule. With just these pieces of information (3D coordinates + normal modes), is it possible to generate a publication-quality figure demonstrating the normal modes?
Of course! The procedure is summarized as follows.
Software requirement
- (Required) VMD version 1.9.1 or later. Freely available: https://www.ks.uiuc.edu/Research/vmd/
- (Optional) Openbabel or other open-source packages that convert xyz files into PDB files.
Overview of the procedure
The open-source visualizer, VMD, has a plugin called NMWizard, which can generate the arrow-annotated molecule structure graphs, and animations of the vibration along the provided normal modes. However, NMWizard cannot directly recognize simple formats like xyz. It requires molecule coordinates to be provided in PDB file format, and normal modes to be provided in .nmd file format. We will talk about how to prepare these files in our example.
Try and learn
1. File preparation
In this example, we are going to look at the normal modes of a CO$_2$ molecule. We are going to use the record of CCSD(T)/cc-pVTZ calculated equilibrium structure and normal modes of CO$_2$ from NIST computational chemistry benchmark dataset. This webpage directly shows the equilibrium coordinates and normal mode vectors in tables.
1.1 Coordinate file (PDB) generation
We first want to prepare the PDB file of the equilibrium structure. Direct preparation of PDB file from scratch is not super efficient unless you already have some script to do so. The reason is that PDB file format has strict requirements for the width of each column and is sensitive to the number of spaces. If you want to write a PDB file for a molecule from scratch with only element and coordinates available, you will need to refer to the PDB format documentation in this link and carefully follow all the requirements.
To avoid errors in writing PDB files, I always first write an xyz file, and then convert it to PDB. An xyz file has extremely simple format requirements
- The first line is the integer indicating the total number of atoms in the molecule
- The second line is for comments. You can leave it blank
- The rest N lines are coordinates of the N atoms. Each row has 4 columns: element symbol, x, y, and z.
- The best thing about
xyzfile format: it has no restrictions on the width of columns, neither do they care about how many spaces you put between two columns. It’s very friendly for manual file preparations.
Following is CO2.xyz:
3
C 0.00000 0.00000 0.00000
O 0.00000 0.00000 1.16637
O 0.00000 0.00000 -1.1663Once you have the xyz file, you can convert it to PDB in different ways. Openbabel is a great open-source chemistry file format converter. However, if you don’t have to install another package on your computer, you can just convert it with VMD. Simplify open VMD, load your xyz file, and right-click the just loaded molecule and select Save coordinates. Then you can choose to save it as PDB.
Following is CO2.PDB I got by converting with VMD. The PDB file converted by Openbabel will look slightly different. However, it doesn’t matter for the purpose of visualizing normal modes.
CRYST1 0.000 0.000 0.000 90.00 90.00 90.00 P 1 1
ATOM 1 C X 1 0.000 0.000 0.000 0.00 0.00 C
ATOM 2 O X 1 0.000 0.000 1.166 0.00 0.00 O
ATOM 3 O X 1 0.000 0.000 -1.166 0.00 0.00 O
ENDAttention This is not a standard PDB file, because, in standard PDB files, each atom in the same residue needs to have a unique atom name (column 3 of this file). To make sure NMWizard correctly recognizes different atoms, I manually edit the names of the two O atoms to follow that standard. Then I have the updated CO2.PDB file as:
CRYST1 0.000 0.000 0.000 90.00 90.00 90.00 P 1 1
ATOM 1 C X 1 0.000 0.000 0.000 0.00 0.00 C
ATOM 2 O1 X 1 0.000 0.000 1.166 0.00 0.00 O
ATOM 3 O2 X 1 0.000 0.000 -1.166 0.00 0.00 O
ENDNow we have the PDB file ready.
1.2 Normal modes file (.nmd) generation
The normal modes file contains the coordinates and some redundant structure information (residue name, atom names, residue number, coordinates of atoms) already included in PDB just to make sure VMD can correctly map the normal modes to the coordinates of different atoms.
Please follow the guideline in the following link to prepare the .nmd file. I wrote some Python scripts to write .nmd file to avoid potential human errors.
Following is the .nmd file I generated for the 4 normal modes of CO$_2$ given by the NIST database.
title CO2
names C O1 O2
resnames X X X
resnums 1 1 1
coordinates 0.00000 0.00000 0.00000 0.00000 0.00000 1.16637 0.00000 0.00000 -1.1663
mode 1 0.00 0.00 0.00 0.00 0.00 0.71 0.00 0.00 -0.71
mode 2 0.00 0.00 0.88 0.00 0.00 -0.33 0.00 0.00 -0.33
mode 3 0.88 0.00 0.00 -0.33 0.00 0.00 -0.33 0.00 0.00
mode 4 0.00 0.88 0.00 0.00 -0.33 0.00 0.00 -0.33 0.002. Load into VMD and visualize normal modes
This part is mainly about clicking through different options of VMD. To avoid confusion, I made a video. Please watch and follow me to visualize the normal modes.
